

Disordered regions (DRs) are defined
as entire proteins or regions of proteins that lack a fixed tertiary structure.
The definition of "disorder" as used here applies to the protein backbone
instead of the residue side-chains. Take a set of proteins, each member the
same. Within this set, the backbones of
ordered protein
regions have the
same Ramachandran angles among the different copies of the protein, while
disordered protein regions
would have different, often dynamic,
Ramachandran angles among the set members.
These DRs are divided into two major classes: Extended (i.e., random coil like). The backbones of these proteins appears to be highly extended, similar to beta sheets. That is, the backbones are stretched out (like a beta sheet's backbone as compared to an alpha-helix's backbone). However, these DRs lack long range contacts which are present in beta sheets. Therefore, the backbone angles fluctuate rapidly, unlike beta sheets.
Collapsed (i.e., molten globule like). These DRs include, but are not limited to, molten globular regions. In general, molten globules have some proportion of secondary structure, but do not have stable tertiary structure. In comparison, collapsed DRs are more general than this, in that secondary structure is not required.
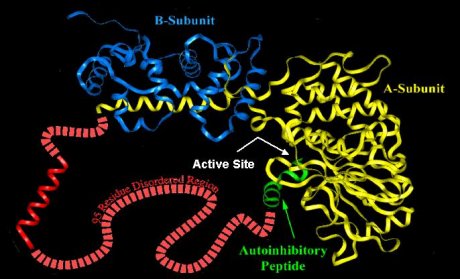
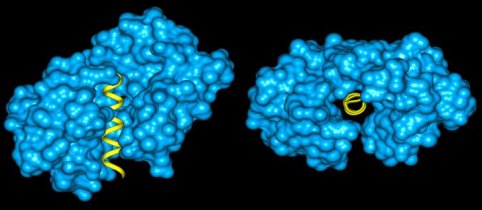
Calcineurin is an example of extended disorder. It is a calcium- and calmodulin-dependent protein serine/threonine phosphatase. It is necessary for several important processes, including T-cell activation. It is composed of two subunits (colored yellow and blue in the image). It has several DRs, including one long stretch of 95 residues (colored red). In that region is the calmodulin binding helix (the red helix), a region of "potential" order. In order for calcineurin to become active, a second protein, calmodulin, must bind to calcineurin. The calmodulin binding site is the stretch of amino acids which form the helix shown above. The DR surrounding (and including) the binding helix allows calmodulin to access that stretch of amino acids and bind to it, wrapping around the helix (see below). It is at this point that the calcineurin disordered region undergoes a disorder-to-order transition. For a further discussion, please see Romero et al., 1997.
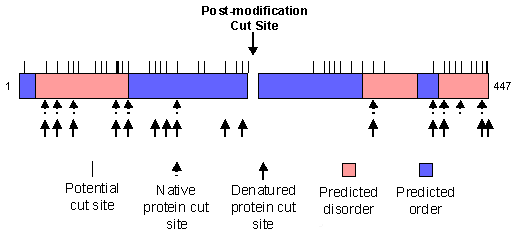
Calmodulin (blue) bound to the helix of calcineurin (yellow). Clusterin is an example of collapsed disorder. It is a protein that was first found in secretions from Sertoli cells (which "nurse" developing germ spells in the testes) and was later found in other tissues, especially the brain. It has been identified as playing a role in apoptosis, cellular injury and lipid transport (Bailey and Griswold, 1999). When clusterin was digested with trypsin, only a portion of the potential cut sites were actually digested. Of the observed cut sites, the large majority were within regions of predicted disorder. When clusterin was denatured, several other digestion sites appeared, all within predicted disordered regions. This is shown graphically, below. Intrinsically disordered regions have been shown to be involved in a variety of functions (see
Dunker et al., 2002 where over 90 examples are listed), including the following: This list is not exhaustive and will expand as more functions are discovered for disordered regions.
Though DRs lack a defined 3-D structure in isolation, many undergo disorder-to-order transitions upon binding to their protein or nucleic acid partners (Dunker et al. 2001, Dyson and Wright 2002).
![]()
![]()
Extended disorder example:
Calcineurin
Collapsed disorder example:
Clusterin
Significance of Disordered Proteins
![]()
DNA/RNA/protein recognition
![]()
Modulation of specificity/affinity of protein binding
![]()
Molecular threading
![]()
Activation by cleavage
Given that amino acid sequence determines 3-D structure, the hypothesis was proposed that sequence would determine lack of structure as well. If local sequence indeed codes for lack of 3-D structure, disordered regions are distinct from structural elements that have been removed from their folding context. To test this, we developed a series of neural network
predictors (NNPs) that use primary structure to make local predictions of disorder. In fact, from the high prediction accuracy achieved, we conclude amino acid sequence does code for DRs. Eight of these Predictors of Natural Disordered Regions (PONDRs) have been developed so far, but PONDR® VL-XT is the most mature of these.
Bailey R and Griswold MD. (1999) Clusterin int he male reproductive system: localization and possible function, Mol. Cell Endocrinol., 151, 17-23.
Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, and Obradovic Z. (2002) Intrinsic disorder and protein function, Biochemistry, 41, 6573-6582.
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, and Obradovic Z. (2001) Intrinsically disordered protein, J. Mol. Graphics and Modeling, 19, 26-59.
Dyson HJ and Wright PE. (2002) Coupling of folding and binding for unstructured proteins, Curr. Opin. Struct. Biol., 12, 54-60.
Romero P, Obradovic Z, and Dunker AK. (1997) Sequence data analysis for long disordered regions prediction in the calcineurin family, Genome Informatics, 8, 110-124.
| ||||||||
Copyright © 2002, 2003 Molecular Kinetics, Inc., all rights reserved.
Access to PONDR® is provided by Molecular Kinetics (6201 La Pas Trail - Ste 160, Indianapolis, IN 46268;
www.molecularkinetics.com;
main@molecularkinetics.com) under license from the WSU Research Foundation.
PONDR® is copyright ©1999 by the WSU Research Foundation, all rights reserved.
Molecular Kinetics, Inc., Washington State University and the WSU Research Foundation and their several employees and consultants assume no liability, either real or implied, from the use of PONDR® in any of its forms or the results of its predictions for any damage, loss of time, loss of profit, either real or potential, or any other damage or loss that may arise from the use of PONDR® in any of its forms or from the results of its predictions. Last updated: Jan. 12, 2007.
