
There are several general guidelines one may use in interpreting PONDR® outputs. Among these are the following.
Look for long regions of prediction. For instance, regions of 30 or more consecutively predicted disordered residues are more
more likely to be a true prediction than regions of 5 consecutively predicted disordered residues.
For example, in RluC the region from residues 34 - 87 is more accurately predicted than the region from 189 - 192.
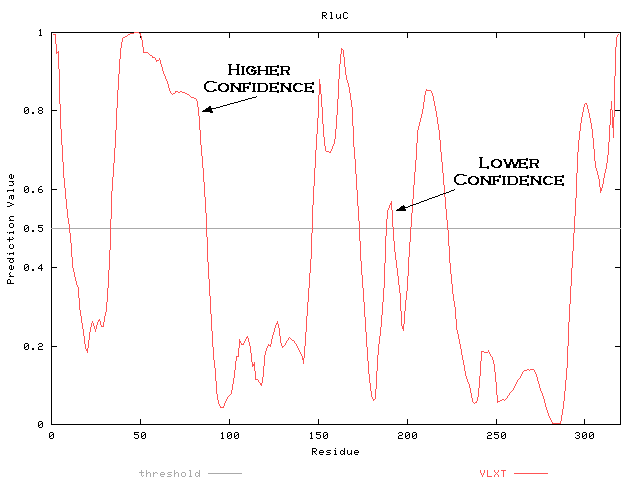
Look for short predictions of order within long predictions of disorder. Also look for regions known to be disorderd, but predicted to be ordered (Garner et al., 1999). These could indicate binding regions or other functions.
For instance, in 4EBP1, a completely disordered protein by NMR analysis, PONDR® predicts a region of order in the center. This region is very close to the eTF4E binding region (the black box in the figure below; see Dunker et al., 2001).
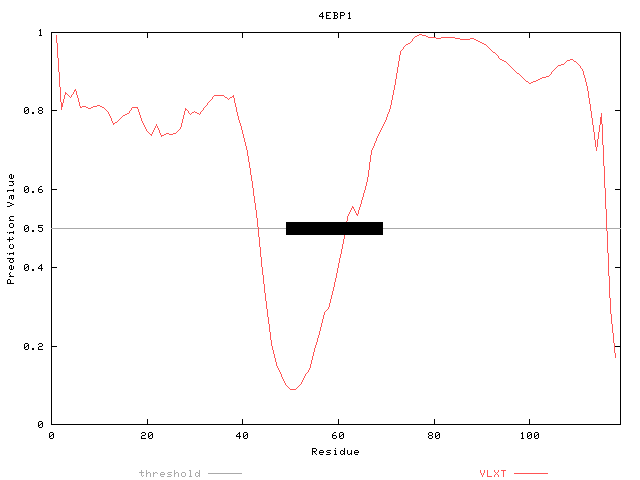
PONDR® VL-XT prediction upon 4EBP1.
The black rectangle on the 0.5 line indicates the region that was visible in the crystal structure with this protein bound to its binding partner, eTF4E.
Look for regions predicted to be disordered, but are shown as ordered in other databases (such as PDB). These could indicate regions of disorder-to-order transition upon some change, such as binding to a partner.
For instance, the p53 oligomerization region
discussed earlier or the c-fos/c-jun/DNA junction complex (PDB entry 1FOS) (see figure below)
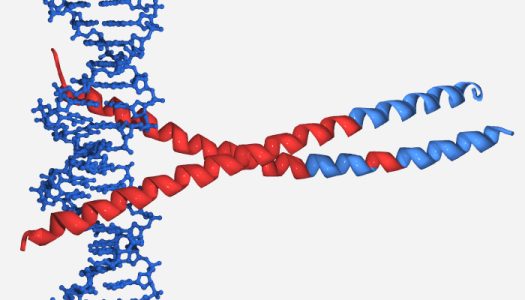
Structure of the c-fos/c-jun/DNA complex.
Only one of the c-fos/c-jun interaction pairs is shown here. PONDR® predicted disorder and order is represented by red and blue ribbons, respectively. Note that the regions of both proteins which interact with DNA is predicted to be disordered.
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Aussio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, and Obradovic Z. (2001) Intrinsically disordered protein, J. Mol. Graphics and Modeling, 19, 26-59.
Garner E, Romero P, Dunker AK, Brown C, and Obradovic Z. (1999) Predicting binding regions within disordered proteins, Genome Informatics, 10, 41-50.
|
Copyright © 2002, 2003 Molecular Kinetics, Inc., all rights reserved.
Access to PONDR® is provided by Molecular Kinetics (6201 La Pas Trail - Ste 160, Indianapolis, IN 46268;
www.molecularkinetics.com;
main@molecularkinetics.com) under license from the WSU Research Foundation.
PONDR® is copyright ©1999 by the WSU Research Foundation, all rights reserved.
Molecular Kinetics, Inc., Washington State University and the WSU Research Foundation and their several employees and consultants assume no liability, either real or implied, from the use of PONDR® in any of its forms or the results of its predictions for any damage, loss of time, loss of profit, either real or potential, or any other damage or loss that may arise from the use of PONDR® in any of its forms or from the results of its predictions. Last updated: Jan. 12, 2007.
