
The PONDR® output consists of up to three parts, depending on what is chosen by the user on the input form:
A graphical representation of the PONDR® output
A textual output using "D" to show disorder along the sequence
A raw score for each predictor for each amino acid in the protein
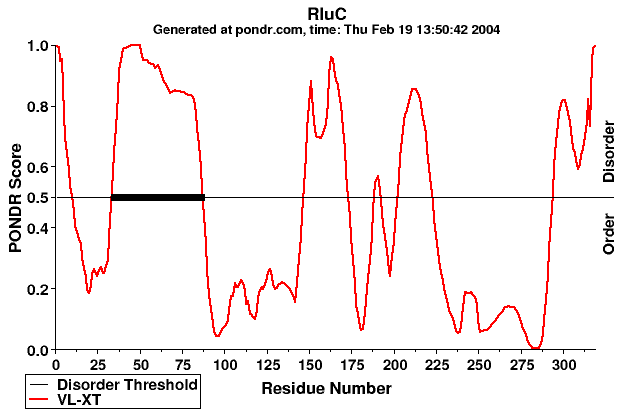
This is the graphical output from a PONDR® prediction, using only the
VL-XT algorithm. The protein sequence input was RluC (SwissProt accession code rluc_ecoli). RluC is the Ribosomal large subunit pseudouridine synthase C (also known as "Pseudouridylate synthase" or "Uracil hydrolyase"). The accession code was entered in the appropriate box in the
input form and then the "Submit" button was pressed.
You may click on various portions of this image to open a small window containing a brief explanation.
|
Copyright © 2002, 2003 Molecular Kinetics, Inc., all rights reserved.
Access to PONDR® is provided by Molecular Kinetics (6201 La Pas Trail - Ste 160, Indianapolis, IN 46268;
www.molecularkinetics.com;
main@molecularkinetics.com) under license from the WSU Research Foundation.
PONDR® is copyright ©1999 by the WSU Research Foundation, all rights reserved.
Molecular Kinetics, Inc., Washington State University and the WSU Research Foundation and their several employees and consultants assume no liability, either real or implied, from the use of PONDR® in any of its forms or the results of its predictions for any damage, loss of time, loss of profit, either real or potential, or any other damage or loss that may arise from the use of PONDR® in any of its forms or from the results of its predictions. Last updated: Jan. 12, 2007.
